Each year, microorganisms containing a certain enzyme remove an estimated 100 million tons of the pollutant carbon monoxide (CO) from the environment. Now, MIT researchers have new insights into how they go about it—happy news for inorganic chemists who have long been trying to synthesize compounds that can do the same thing without the living creature.
“Microorganisms such as bacteria can do lots of chemistry that people would like to do,” says Catherine L. Drennan, professor of chemistry and a Howard Hughes Medical Institute investigator. “They can form and break carbon bonds, split nitrogen, and break apart hydrogen and oxygen—all things that we can’t do or can do only with great difficulty.”
Key to the microorganisms’ ability to perform such feats are large, powerful enzymes that catalyze (speed up) reactions. For many decades, researchers worldwide have been working to replicate that chemistry using smaller molecules in an artificial rather than natural setting. Success could mean the ability to make hydrogen for fuel cells, to remove the greenhouse gas carbon dioxide (CO2) from the atmosphere, to clean up CO in polluted urban areas, and more.
But the researchers’ best efforts are frequently unsuccessful—and Drennan is not surprised. “If we’re going to copy what microorganisms are doing, we need to have a clear understanding of how they do it—at the molecular level,” she says. She and her research group aim to develop that understanding by actually observing the physical structure of the key molecules involved and seeing how they change when a reaction takes place.
Recently, her work has focused on an enzyme that—depending on its changeable structure—can take up CO and release CO2, or take up CO2 and release CO, or use the CO to make a form of acetate that plays a key role in metabolism. “Unlike humans, these organisms are very flexible,” says Drennan. “They’ll take whatever is around them and find a way to live on it.”
To start, she has been investigating the reaction whereby CO is picked up by the enzyme, where it reacts with water to form CO2. That chemistry can be traced to a small section within the enzyme known as the C-cluster—an unusual and possibly quite ancient combination of metals and inorganic compounds including iron, nickel, and sulfur.
To help inorganic chemists replicate the abilities of the C-cluster, Drennan has been exploring its structural details. Using crystalline enzyme samples, she has looked at where the atoms are located, how they are oriented, and where empty sites are needed for other atoms to attach and catalyze chemical reactions.
Using X-rays to “see” atoms
Atoms are too small to see with an optical microscope, so Drennan turns to X-rays, which have a wavelength a thousand times shorter than that of visible light and comparable to the spacing of atoms in a crystal. The technique she uses, called X-ray crystallography, involves beaming X-rays through a crystal sample. Atoms in the sample diffract the X-rays, creating a diffraction pattern that a crystallographer—with the help of mathematical methods—converts into an electron density map and ultimately to an image, or “snapshot,” that shows where the atoms in the sample are located.
For these studies, Drennan receives samples of the enzyme from collaborator Stephen Ragsdale at the University of Michigan Medical School, who has a laboratory specially equipped to grow the microorganism of interest. The microorganisms grow rapidly at room temperature, and the enzyme is abundant and stable—except that the metal clusters are sensitive to oxygen, so all work takes place in chambers filled with argon or nitrogen.
Keeping their enzymes away from oxygen is a minor inconvenience compared with the challenges involved in using X-ray crystallography to study them. First, the researchers must get the enzyme to form a crystal—a task that Drennan deems the “hard part,” which can take many years of trying different materials and methods. In this case, her team uses salt at high concentrations to force the enzyme molecules out of solution and into a crystalline state. The individual enzymes line up in a regularly repeating pattern in a three-dimensional crystal, presenting enough sample to be analyzed.
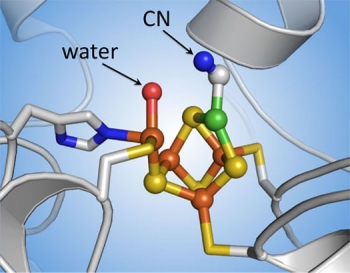
The next challenge is to stop the reaction just before it happens. “With X-ray crystallography you only get snapshots in time,” says Drennan. “If you give the enzyme everything it needs for the reaction, it’ll react—and your snapshot will show the end result but not how it happened.” Tests showed that providing the enzyme with cyanide (CN) rather than CO does the trick. CN is similar to CO in structure and will bind at the same site where the CO would bind—but it won’t react. The enzyme will be poised for action but frozen in time.
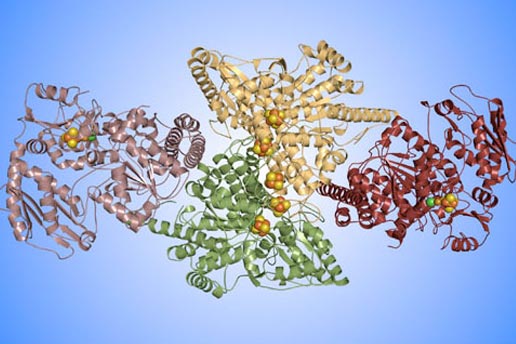
This diagram shows the overall structure of an enzyme that enables microorganisms to capture large quantities of carbon dioxide and carbon monoxide from the environment. Using X-ray crystallography, MIT researchers have solved the enzyme’s three-dimensional structure to atomic resolution. The colors in this “ribbon” diagram indicate the four protein chains that make up the enzyme. The grouped spheres are clusters of metal atoms that play key roles in the carbon chemistry. Diagram: Yan Kung, MIT
Atomic-level insights
Their experiments worked well. They now have a clear image of the arrangement of the metal atoms in the C-cluster just prior to the reaction. They can see where the CO would bind, and they know the location of the nearby water molecule that participates in the reaction. From those observations, they can predict how the reaction would proceed.
The researchers’ results settled a long-standing debate about the presence or absence of a single sulfur atom. Another group has argued that there is a sulfur atom at the site where the CN latched on. But Drennan’s results suggest that if a sulfur atom were there, it would block the CO from binding. Most experts in the field now agree that the “active” form of the C-cluster has no sulfur in that position—a finding significant for inorganic chemists as they manipulate materials to mimic the cluster’s CO-removal action.
Results from Holger Dobbek and his research group at the Max-Planck-Institut für Biochemie (Germany) both verify and supplement the MIT findings. That group produced an image of the C-cluster with a CO2 molecule in place—a structure that can be interpreted as a post-reaction counterpart to Drennan’s pre-reaction images with CN (the stand-in for CO). Indeed, the structures from the two groups superimpose remarkably well, and in neither case is the disputed sulfur atom in evidence. “So at an atomic resolution, we have images that enable us to understand one of the important chemical reactions that happens on this metal site,” says Drennan. “It’s really very exciting.”
Drennan and her colleagues are now investigating other metal clusters in the same enzyme, in particular, one that controls the acetate-forming reactions. But, she warns, there is always the chance that—even getting the right structure—human-made copies of the metal clusters may not work. For example, it may be impossible to make a small version that is stable but still flexible enough to do chemistry. (In the CO reaction, for example, the carbon atom needs to rotate to react with the water.) Or the reaction may require other elements in the enzyme, not just the metals. As a result, it may be necessary to use the whole enzyme or perhaps even the whole microorganism to achieve the desired effect.
From a commercial perspective, this particular enzyme is attractive because it can be made in large quantities and at room temperature. The only downside is having to keep it away from oxygen—a problem Drennan thinks she can fix. “I think I know the source of the problem with oxygen,” she says. “We may be able to redesign the enzyme to make it more stable in an oxygen environment.”
This research was funded by the National Institutes of Health and by a seed grant from the MIT Energy Initiative. More information can be found in:
Y. Kung, T. Doukov, J. Seravalli, S. Ragsdale, and C. Drennan. “Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.” Biochemistry, vol. 48, no. 31, 2009, pp. 7432–7440.
This article appears in the Autumn 2009 issue of Energy Futures.